Gosling: A Grammar-based Toolkit for Scalable and Interactive Genomics Data Visualization
Sehi L'Yi, Qianwen Wang, Fritz Lekschas, Nils Gehlenborg
External link (DOI)
View presentation:2021-10-28T15:30:00ZGMT-0600Change your timezone on the schedule page
2021-10-28T15:30:00Z
Fast forward
Direct link to video on YouTube: https://youtu.be/Wvyb9dus9eU
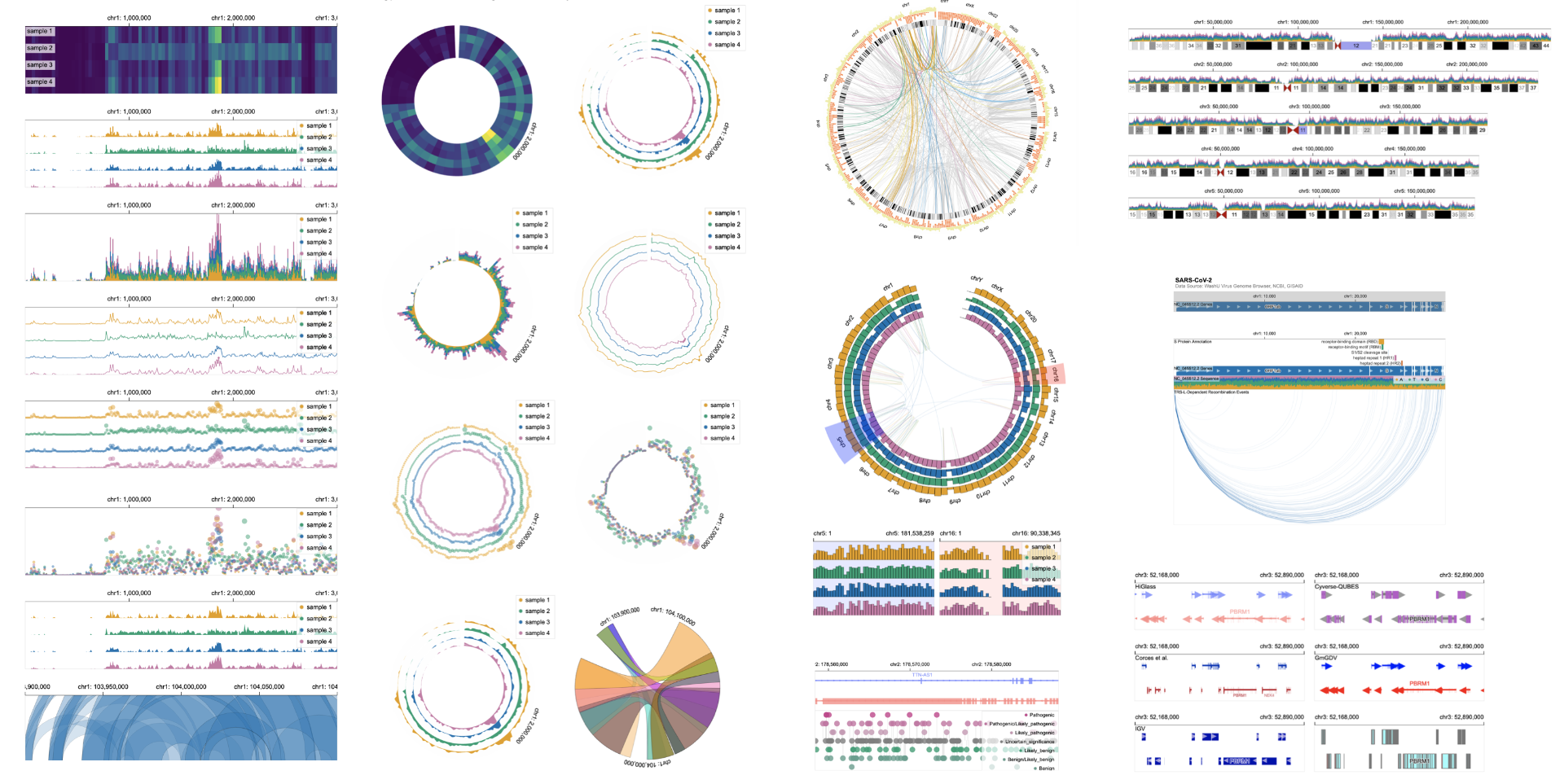
Abstract
The combination of diverse data types and analysis tasks in genomics has resulted in the development of a wide range of visualization techniques and tools. However, most existing tools are tailored to a specific problem or data type and offer limited customization, making it challenging to optimize visualizations for new analysis tasks or datasets. To address this challenge, we designed Gosling—a grammar for interactive and scalable genomics data visualization. Gosling balances expressiveness for comprehensive multi-scale genomics data visualizations with accessibility for domain scientists. Our accompanying JavaScript toolkit called Gosling.js provides scalable and interactive rendering. Gosling.js is built on top of an existing platform for web-based genomics data visualization to further simplify the visualization of common genomics data formats. We demonstrate the expressiveness of the grammar through a variety of real-world examples. Furthermore, we show how Gosling supports the design of novel genomics visualizations. An online editor and examples of Gosling.js, its source code, and documentation are available at https://gosling.js.org.