ChemVA: Interactive Visual Analysis of Chemical Compound Similarity in Virtual Screening
María Virginia Sabando, Pavol Ulbrich, Matias Selzer, Jan Byška, Jan Mičan, Ignacio Ponzoni, Axel Soto, María Luján Ganuza, Barbora Kozlikova
External link (DOI)
View presentation:2020-10-28T16:45:00ZGMT-0600Change your timezone on the schedule page
2020-10-28T16:45:00Z
Fast forward
Direct link to video on YouTube: https://youtu.be/vKMRGer-pAY
Keywords
Virtual screening, visual analysis, dimensionality reduction, coordinated views, cheminformatics.
Abstract
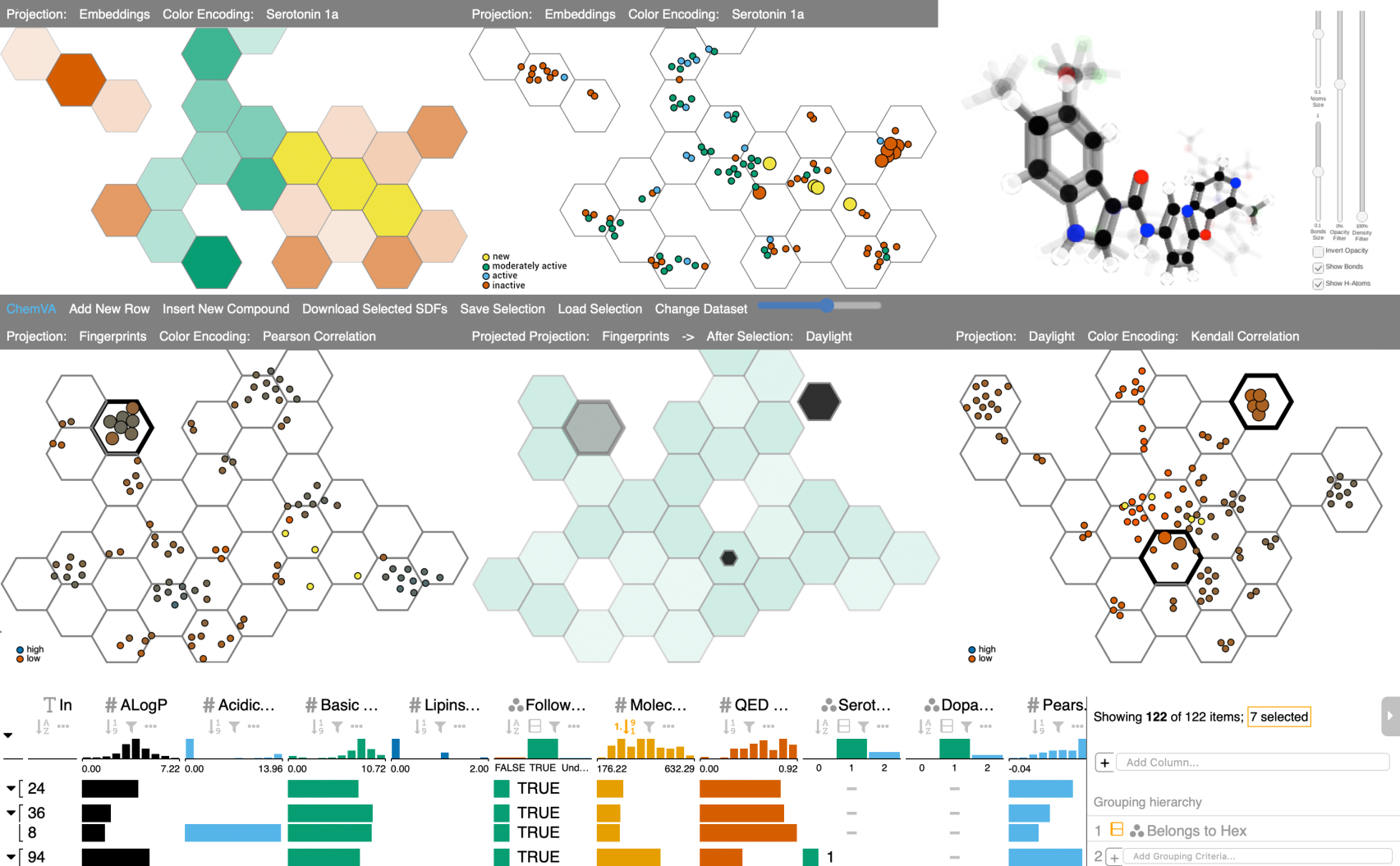
In the modern drug discovery process, medicinal chemists deal with the complexity of analysis of large ensembles of candidate molecules. Computational tools, such as dimensionality reduction (DR) and classification, are commonly used to efficiently process the multidimensional space of features. These underlying calculations often hinder interpretability of results and prevent experts from assessing the impact of individual molecular features on the resulting representations. To provide a solution for scrutinizing such complex data, we introduce ChemVA, an interactive application for the visual exploration of large molecular ensembles and their features. Our tool consists of multiple coordinated views: Hexagonal view, Detail view, 3D view, Table view, and a newly proposed Difference view designed for the comparison of DR projections. These views display DR projections combined with biological activity, selected molecular features, and confidence scores for each of these projections. This conjunction of views allows the user to drill down through the dataset and to efficiently select candidate compounds. Our approach was evaluated on two case studies of finding structurally similar ligands with similar binding affinity to a target protein, as well as on an external qualitative evaluation. The results suggest that our system allows effective visual inspection and comparison of different high-dimensional molecular representations. Furthermore, ChemVA assists in the identification of candidate compounds while providing information on the certainty behind different molecular representations.