ProtEGOnist – Exploration of protein-protein interactions using ego-graph networks
Nicolas Brich, Theresa Harbig, Mathias Witte Paz, Simon Tim Hackl, Caroline Jachmann, Marco Schäfer, Michael Krone, Kay Nieselt
Room: 111-112
2023-10-22T22:00:00ZGMT-0600Change your timezone on the schedule page
2023-10-22T22:00:00Z
Fast forward
Abstract
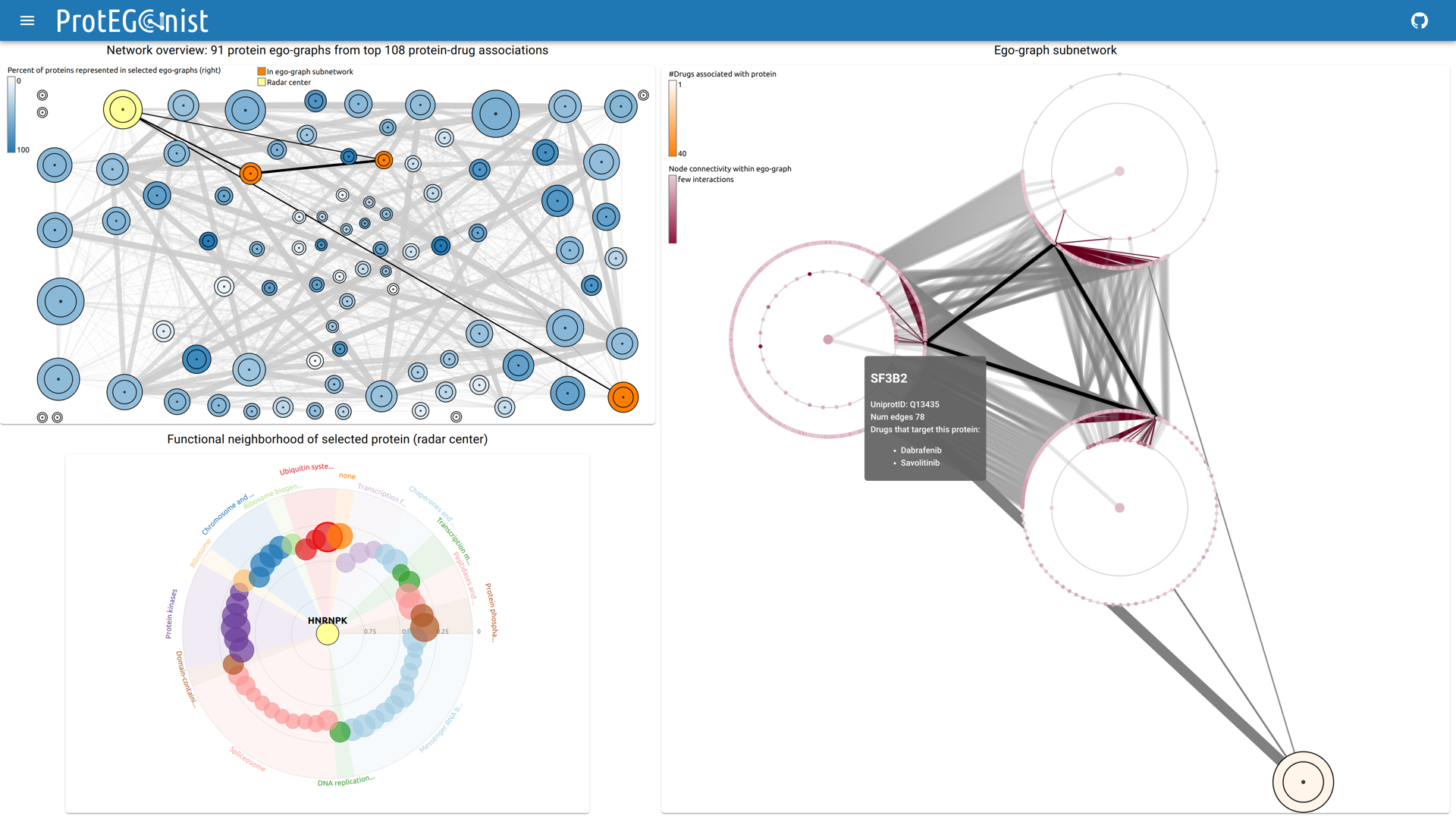
The complexity of protein-protein interaction (PPI) networks often leads to visual clutter and limited interpretability. To overcome these problems, we present ProtEGOnist, a novel, interactive visualization approach designed to explore PPI networks with a focus on drug-protein associations. ProtEGOnist addresses the challenges by introducing the concept of ego-graphs to represent local PPI neighborhoods around proteins of interest. These ego-graphs are aggregated into an ego-graph network, where edges between ego-graphs encoded their similarity using the Jaccard index. Our proposed visualization design offers an overview of drug-associated proteins, radar charts to compare protein functions, and detailed ego-graph subnetworks for interactive exploration. Our aim was to reduce visual complexity while enabling detailed exploration, facilitating the discovery of meaningful patterns in PPI networks. A web-based prototype of ProtEGOnist is available for interactive use.