Vimo: Visual Analysis of Neuronal Connectivity Motifs
Jakob Troidl, Simon Alexander Warchol, Jinhan Choi, Jordan Matelsky, Nagaraju Dhanyasi, Xueying Wang, Brock Wester, Donglai Wei, Jeff Lichtman, Hanspeter Pfister, Johanna Beyer
DOI: 10.1109/TVCG.2023.3327388
Room: 103
2023-10-25T22:48:00ZGMT-0600Change your timezone on the schedule page
2023-10-25T22:48:00Z
Fast forward
Full Video
Keywords
Visual motif analysis, Focus&Context, Scientific visualization, Neuroscience, Connectomics.
Abstract
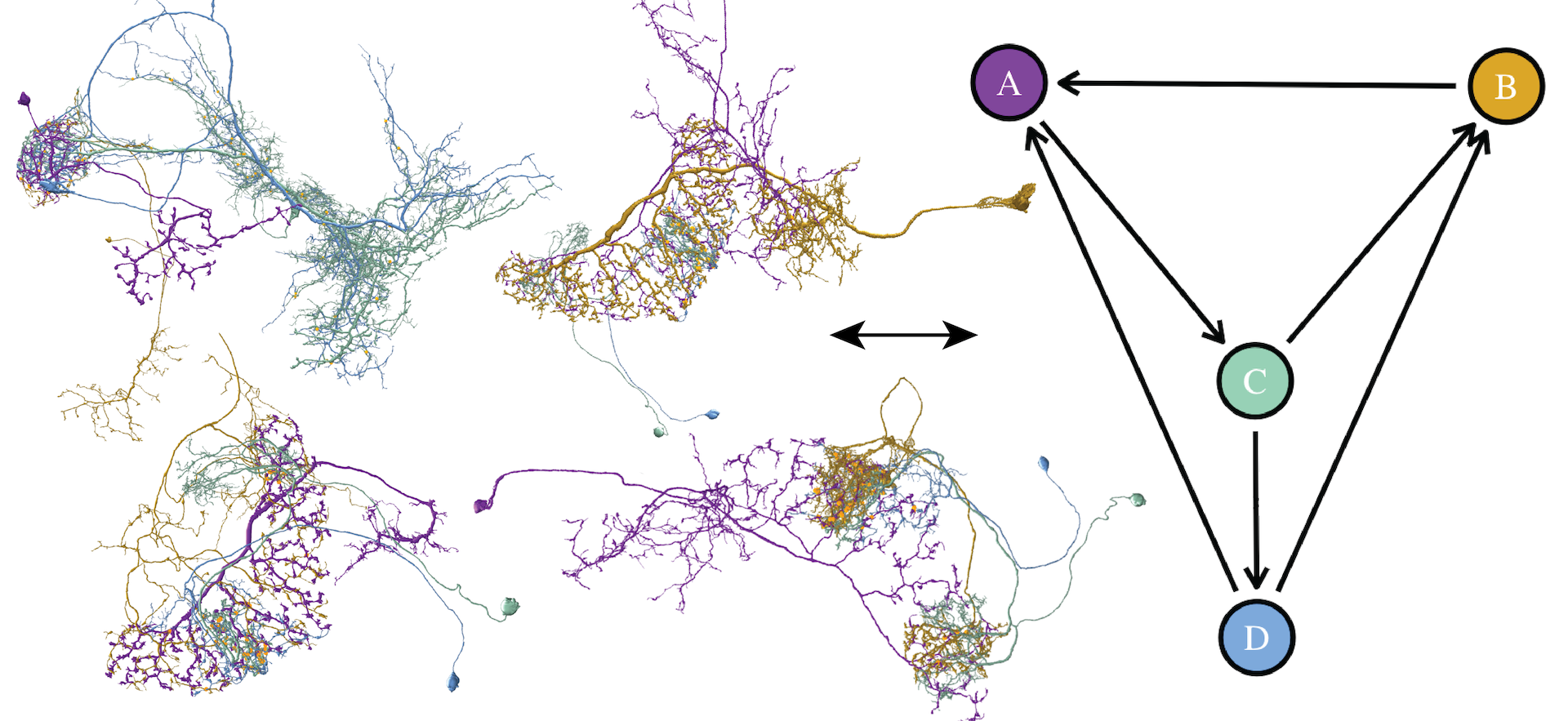
Recent advances in high-resolution connectomics provide researchers with access to accurate petascale reconstructions of neuronal circuits and brain networks for the first time. Neuroscientists are analyzing these networks to better understand information processing in the brain. In particular, scientists are interested in identifying specific small network motifs, i.e., repeating subgraphs of the larger brain network that are believed to be neuronal building blocks. Although such motifs are typically small (e.g., 2 -6 neurons), the vast data sizes and intricate data complexity present significant challenges to the search and analysis process. To analyze these motifs, it is crucial to review instances of a motif in the brain network and then map the graph structure to detailed 3D reconstructions of the involved neurons and synapses. We present Vimo, an interactive visual approach to analyze neuronal motifs and motif chains in large brain networks. Experts can sketch network motifs intuitively in a visual interface and specify structural properties of the involved neurons and synapses to query large connectomics datasets. Motif instances (MIs) can be explored in high-resolution 3D renderings. To simplify the analysis of MIs, we designed a continuous focus&context metaphor inspired by visual abstractions. This allows users to transition from a highly-detailed rendering of the anatomical structure to views that emphasize the underlying motif structure and synaptic connectivity. Furthermore, Vimo supports the identification of motif chains where a motif is used repeatedly (e.g., 2 -4 times) to form a larger network structure. We evaluate Vimo in a user study and an in-depth case study with seven domain experts on motifs in a large connectome of the fruit fly, including more than 21,000 neurons and 20 million synapses. We find that Vimo enables hypothesis generation and confirmation through fast analysis iterations and connectivity highlighting.