A simple pipeline for creating cover quality images of post-translational modifications to molecular structures in rare disease
Mo Rahman, Onno Faber, Pascale Marill, Otavio Good
View presentation:2022-10-16T19:40:00ZGMT-0600Change your timezone on the schedule page
2022-10-16T19:40:00Z
The live footage of the talk, including the Q&A, can be viewed on the session page, Bio+MedVis: Challenges.
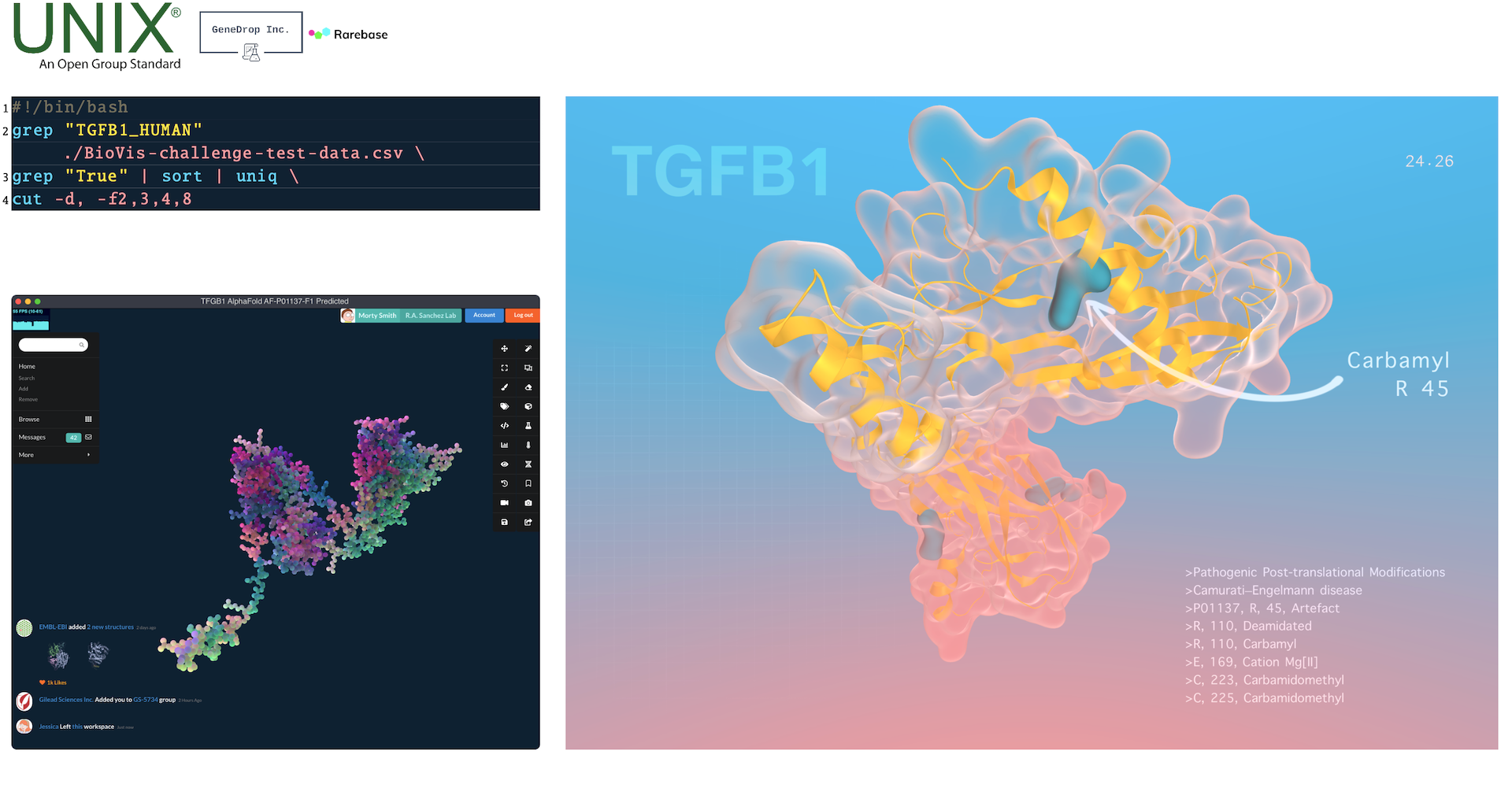
Abstract
Changes to molecular structure can have a large impact on biological function and are the basis for many diseases. Structural modifications to proteins, while not changes to the amino acid sequence, can alter their function and have been implicated in rare diseases. Visualizing these modifications on a 3D model helps understand how the characteristics of a molecule are affected. We present a simple pipeline to create high-quality rendered images of post-translational protein modifications. Data provided by CompOmics group at VIB and Ghent University in Ghent, Belgium is parsed using standard UNIX commands, then added to AlphaFold predicted structures using a newly developed viewer Dalton, and finally rendered to an image. Well-designed and simple to make visualizations demystify rare diseases and mechanisms of post-translational structural modifications, making it easier to conduct further research in these fields.