NeuRegenerate: A Framework for Visualizing Neurodegeneration
Saeed Boorboor, Shawn Mathew, Mala Ananth, David Talmage, Lorna W. Role, Arie E. Kaufman.
View presentation:2022-10-20T14:48:00ZGMT-0600Change your timezone on the schedule page
2022-10-20T14:48:00Z
Prerecorded Talk
The live footage of the talk, including the Q&A, can be viewed on the session page, Neuro/Brain/Medical Data.
Fast forward
Keywords
Neuron visualization, volume visualization, volume transformation, neuroscience, wide-field microscopy, machine learning,
Abstract
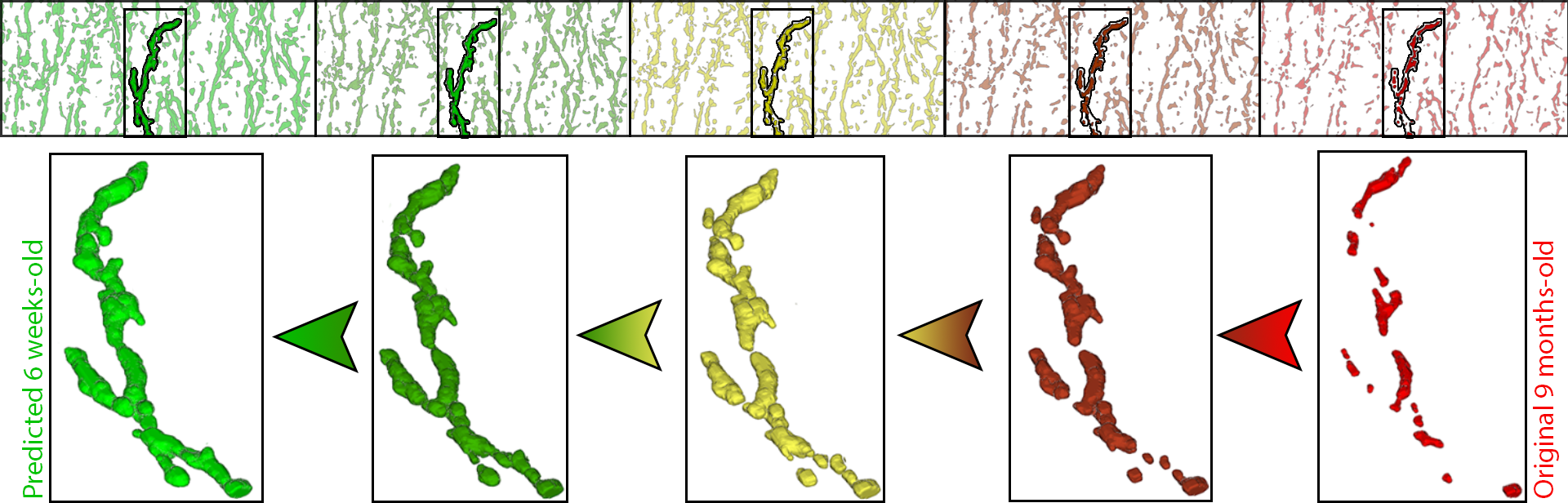
Recent advances in high-resolution microscopy have allowed scientists to better understand the underlying brain connectivity. However, due to the limitation that biological specimens can only be imaged at a single timepoint, studying changes to neural projections over time is limited to observations gathered using population analysis. In this paper, we introduce NeuRegenerate, a novel end-to-end framework for the prediction and visualization of changes in neural fiber morphology within a subject across specified age-timepoints. To predict projections, we present neuReGANerator, a deep-learning network based on cycle-consistent generative adversarial network (GAN) that translates features of neuronal structures across age-timepoints for large brain microscopy volumes. We improve the reconstruction quality of the predicted neuronal structures by implementing a density multiplier and a new loss function, called the hallucination loss. Moreover, to alleviate artifacts that occur due to tiling of large input volumes, we introduce a spatial-consistency module in the training pipeline of neuReGANerator. Finally, to visualize the change in projections, predicted using neuReGANerator, NeuRegenerate offers two modes: (i) neuroCompare to simultaneously visualize the difference in the structures of the neuronal projections, from two age domains (using structural view and bounded view), and (ii) em neuroMorph, a vesselness-based morphing technique to interactively visualize the transformation of the structures from one age-timepoint to the other. Our framework is designed specifically for volumes acquired using wide-field microscopy. We demonstrate our framework by visualizing the structural changes within the cholinergic system of the mouse brain between a young and old specimen.